MyTrueancestry analysis of my ancestry
Analysed DNA
Y DNA: H-M69 -> H-M82 -> SK1225 -> H-Z5888 -> H-Z5890 -> H-CTS8144 [CTS8144/PF1741/M5498] -> Z34531 (H1a1a4b3b1a8~)
was found 2875 BCE -> Jiroft/IVC Periphery 11459 Shahr-i-Sokte BA2
10k YBP South Asian neolithic route has this H-M69 distribution along the coasts of Iran, Arabia, South Asia & South East Asia has bidirectional movement. There were further waves of trading networks from Iran/Arabia/Central Asia and Global Megalithic route covering Europe, West Asia and Japan/Oceania. Close to 420 Million people in South Asia belong to this Haplogroup.
There are admixtures from Armenia, Parthia, Greco-Bactria, Central Asia Steppe after that. The Lactose persistence SNPS rs3213871 rs4988243 rs2236783 rs3769005 rs4988183 points to heavy Steppe/Middle East influence.
G25Ancients nmonte Dist 0.79 IND_Roopkund_A 51.05 IRN_Shahr_I_Sokhta_BA2 46.64 MAR_Iberomaurusian 2.04 PAK_Katelai_IA 0.19 TKM_Gonur2_BA 0.08
Lactose Persistence
I am only person matching Both the lactose persistence genes that this DA125, Kangju, 150-300 AD genome matches in the 1000 genomes DB
rs4988243 2 136607703 Found in Korea, ITU, BEB, GIH, qatar, MiddleEast, finn, jewish, amerind/latinos (ITU=34%, GIH=32.8%)
rs2236783 2 136594158 Found in Europe and among Amerind/Latinos (ITU=11.27% GIH=12.25%)
[gallery ids="172,173" type="rectangular"]
MyTrueancestry analysis of my ancestry
Shows big influence from Scythian/Sarmatian/Cimmeran/Alan & Avar/Hun groups probably indicating some Saka/Hun ancestry mix
There is also some European matches showing in Ancestry due to admix with Kang Ju like Samples that had significant European heritage and Greco-Bactrian samples.
Some of the Interesting samples I get on Timeline include Oetzi, Hittites, Caananites, Minoan, Maykop, Iran/Arm CHL, Eneolithic Steppe, EBA samples, Many Hungarian Conquerors, Sarmatians, Scythians, Cimmeran, Crusaders, French Samples probably pointing to ancient Pallava, Saka, Rashtrakuta, Chauhans, Ikshvakus etc...
Possibly many royal Scotland, England France Spain Hungary etc.. houses harbor these mix and show up on my Gedmatch matches
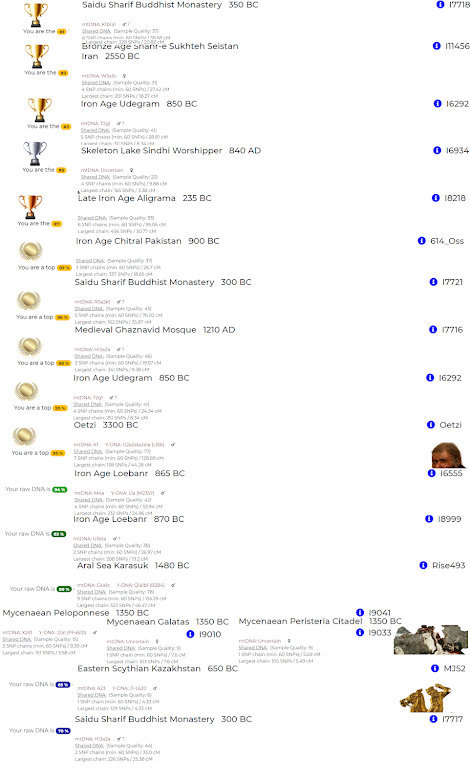
The samples such as I7718, I7717, I7721, I7716 etc.. which show up in the top might be due to the Kamboja influence in Iron Age when ancient Jain group mixed with their Cavalry groups to form Kambojas all over India. Many calculators show at least 5% of this admixture and also up to 15% of Kashmiri groups indicating the high presence of these groups in Kashmir in ancient India. The Oetzi might have been from the ancient maritime trade routes along Megalithic towns present all over the India. Probably few groups in the South harbored more of this autosome in the past since we have evidence of extensive maritime contact during megalithic era in Adichanallur close to 2000BC. The later era saw numerous towns showing evidence of maritime trade with Greeks and Romans.

| |U2’3’4’7’8’9 41500 B.C.E. (Mother: U)
| |U2 41000 B.C.E. (Mother: U2’3’4’7’8’9)
| |U2a 21000 B.C.E. (Mother: U2)
| |U2a1 11500 B.C.E. (Mother: U2a)
| |U2a1a NW South Asia (Mother: U2a1)
-> U2a1a2 (recent Indian mutation same as my mtDNA)
| |U2a1b India,Europe (Mother: U2a1)
| |U2a2 1500 (Mother: U2a)





No comments:
Post a Comment